Cordex domains#
The domain module should give some tools to work with preconfigured or user defined domains. Domains are defined as xarray datasets that will contain dimensions and coodinates according to CF-conventions.
NOTE: The domain module mostly focuses on working with rotated cordex domains and how they are defined in the cordex archive specifications. However, there are some regional models that use different mappings instead of rotated_pole or rotated_latitude_longitude which we focus on. Any expertise working with those different mappings is highly welcome!
Working with domain information#
import cordex as cx
The domain module contains some useful functions to work with cordex meta data, e.g., you can get some domain grid information using
cx.domain_info("EUR-11")
Downloading file 'rotated-latitude-longitude.csv' from 'https://raw.githubusercontent.com/WCRP-CORDEX/domain-tables/main/rotated-latitude-longitude.csv' to '/Users/runner/.py-cordex'.
{'short_name': 'EUR-11',
'region': 4,
'domain_id': 'EUR-12',
'domain': 'Europe',
'CORDEX_domain': 'EUR-11',
'long_name': 'Europe',
'nlon': 424,
'nlat': 412,
'll_lon': -28.375,
'ur_lon': 18.155,
'll_lat': -23.375,
'ur_lat': 21.835,
'dlon': 0.11,
'dlat': 0.11,
'pollon': -162.0,
'pollat': 39.25}
The domain information is stored in a number of csv tables. The module contains a tables dictionary that sorts the tables by resolution or project, e.g.
cx.domains.tables.keys()
Index(['region', 'domain_id', 'domain', 'CORDEX_domain', 'long_name', 'nlon',
'nlat', 'll_lon', 'ur_lon', 'll_lat', 'ur_lat', 'dlon', 'dlat',
'pollon', 'pollat'],
dtype='object')
All available cordex domains are in those tables:
cx.domains.table
| region | domain_id | domain | CORDEX_domain | long_name | nlon | nlat | ll_lon | ur_lon | ll_lat | ur_lat | dlon | dlat | pollon | pollat | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| short_name | |||||||||||||||
| SAM-44 | 1 | SAM-50 | South America | SAM-44 | South America | 146 | 167 | 143.92000 | 207.72000 | -38.28000 | 34.76000 | 0.4400 | 0.4400 | -56.06 | 70.60 |
| CAM-44 | 2 | CAM-50 | Central America | CAM-44 | Central America | 210 | 113 | -52.80000 | 39.16000 | -28.60000 | 20.68000 | 0.4400 | 0.4400 | 113.98 | 75.74 |
| NAM-44 | 3 | NAM-50 | North America | NAM-44 | North America | 155 | 130 | -33.88000 | 33.88000 | -28.40000 | 28.36000 | 0.4400 | 0.4400 | 83.00 | 42.50 |
| EUR-44 | 4 | EUR-50 | Europe | EUR-44 | Europe | 106 | 103 | -28.21000 | 17.99000 | -23.21000 | 21.67000 | 0.4400 | 0.4400 | -162.00 | 39.25 |
| AFR-44 | 5 | AFR-50 | Africa | AFR-44 | Africa | 194 | 201 | -24.64000 | 60.28000 | -45.76000 | 42.24000 | 0.4400 | 0.4400 | 180.00 | 90.00 |
| WAS-44 | 6 | WAS-50 | South Asia | WAS-44 | South Asia | 193 | 130 | -32.12000 | 52.36000 | -21.56000 | 35.20000 | 0.4400 | 0.4400 | -123.34 | 79.95 |
| EAS-44 | 7 | EAS-50 | East Asia | EAS-44 | East Asia | 203 | 167 | -40.92000 | 47.96000 | -26.84000 | 46.20000 | 0.4400 | 0.4400 | -64.78 | 77.61 |
| CAS-44 | 8 | CAS-50 | Central Asia | CAS-44 | Central Asia | 153 | 100 | -34.32000 | 32.56000 | -20.68000 | 22.88000 | 0.4400 | 0.4400 | -103.39 | 43.48 |
| AUS-44 | 9 | AUS-50 | Australasia | AUS-44 | Australasia | 200 | 129 | 142.16000 | 229.72000 | -22.88000 | 33.44000 | 0.4400 | 0.4400 | 141.38 | 60.31 |
| ANT-44 | 10 | ANT-50 | Antarctica | ANT-44 | Antarctica | 125 | 97 | 152.72000 | 207.28000 | -27.72000 | 14.52000 | 0.4400 | 0.4400 | -166.92 | 6.08 |
| ARC-44 | 11 | ARC-50 | Arctic | ARC-44 | Arctic | 116 | 133 | -22.88000 | 27.72000 | -24.20000 | 33.88000 | 0.4400 | 0.4400 | 0.00 | 6.55 |
| MED-44 | 12 | MED-50 | Mediterranean | MED-44 | Mediterranean | 98 | 63 | -23.22000 | 19.46000 | -21.34000 | 5.94000 | 0.4400 | 0.4400 | 198.00 | 39.25 |
| MNA-44 | 13 | MNA-50 | Middle East and North Africa | MNA-44 | Middle East and North Africa | 232 | 118 | -26.40000 | 75.24000 | -6.60000 | 44.88000 | 0.4400 | 0.4400 | 180.00 | 90.00 |
| MNA-22 | 13 | MNA-25 | Middle East and North Africa | MNA-22 | Middle East and North Africa | 464 | 236 | -26.51000 | 75.35000 | -6.71000 | 44.99000 | 0.2200 | 0.2200 | 180.00 | 90.00 |
| SAM-11 | 1 | SAM-12 | South America | SAM-11 | South America | 584 | 668 | 143.75500 | 207.88500 | -38.44500 | 34.92500 | 0.1100 | 0.1100 | -56.06 | 70.60 |
| CAM-11 | 2 | CAM-12 | Central America | CAM-11 | Central America | 840 | 452 | -52.96500 | 39.32500 | -28.76500 | 20.84500 | 0.1100 | 0.1100 | 113.98 | 75.74 |
| NAM-11 | 3 | NAM-12 | North America | NAM-11 | North America | 620 | 520 | -34.04500 | 34.04500 | -28.56500 | 28.52500 | 0.1100 | 0.1100 | 83.00 | 42.50 |
| EUR-11 | 4 | EUR-12 | Europe | EUR-11 | Europe | 424 | 412 | -28.37500 | 18.15500 | -23.37500 | 21.83500 | 0.1100 | 0.1100 | -162.00 | 39.25 |
| AFR-11 | 5 | AFR-12 | Africa | AFR-11 | Africa | 776 | 804 | -24.80500 | 60.44500 | -45.92500 | 42.40500 | 0.1100 | 0.1100 | 180.00 | 90.00 |
| WAS-11 | 6 | WAS-12 | South Asia | WAS-11 | South Asia | 772 | 520 | -32.28500 | 52.52500 | -21.72500 | 35.36500 | 0.1100 | 0.1100 | -123.34 | 79.95 |
| EAS-11 | 7 | EAS-12 | East Asia | EAS-11 | East Asia | 812 | 668 | -41.08500 | 48.12500 | -27.00500 | 46.36500 | 0.1100 | 0.1100 | -64.78 | 77.61 |
| CAS-11 | 8 | CAS-12 | Central Asia | CAS-11 | Central Asia | 612 | 400 | -34.48500 | 32.72500 | -20.84500 | 23.04500 | 0.1100 | 0.1100 | -103.39 | 43.48 |
| AUS-11 | 9 | AUS-12 | Australasia | AUS-11 | Australasia | 800 | 516 | 141.99500 | 229.88500 | -23.04500 | 33.60500 | 0.1100 | 0.1100 | 141.38 | 60.31 |
| ANT-11 | 10 | ANT-12 | Antarctica | ANT-11 | Antarctica | 500 | 388 | 152.55500 | 207.44500 | -27.88500 | 14.68500 | 0.1100 | 0.1100 | -166.92 | 6.08 |
| ARC-11 | 11 | ARC-12 | Arctic | ARC-11 | Arctic | 464 | 532 | -23.04500 | 27.88500 | -24.36500 | 34.04500 | 0.1100 | 0.1100 | 0.00 | 6.55 |
| MED-11 | 12 | MED-12 | Mediterranean | MED-11 | Mediterranean | 392 | 252 | -23.38500 | 19.62500 | -21.50500 | 6.10500 | 0.1100 | 0.1100 | 198.00 | 39.25 |
| MNA-11 | 13 | MNA-12 | Middle East and North Africa | MNA-11 | Middle East and North Africa | 928 | 472 | -26.56500 | 75.40500 | -6.76500 | 45.04500 | 0.1100 | 0.1100 | 180.00 | 90.00 |
| GAR-0275 | 4 | GAR-3 | Greater Alpine Region | GAR-0275 | Greater Alpine Region | 476 | 444 | -13.51125 | -0.44875 | -10.93125 | 1.25125 | 0.0275 | 0.0275 | -162.00 | 39.25 |
| CEU-0275 | 4 | CEU-3 | Central Europe | CEU-0275 | Central Europe | 345 | 385 | -9.30375 | NaN | -5.07375 | NaN | 0.0275 | 0.0275 | -162.00 | 39.25 |
| SAM-22 | 1 | SAM-25 | South America | SAM-22 | South America | 292 | 334 | 143.81000 | 207.83000 | -38.39000 | 34.87000 | 0.2200 | 0.2200 | -56.06 | 70.60 |
| CAM-22 | 2 | CAM-25 | Central America | CAM-22 | Central America | 420 | 226 | -52.91000 | 39.27000 | -28.71000 | 20.79000 | 0.2200 | 0.2200 | 113.98 | 75.74 |
| NAM-22 | 3 | NAM-25 | North America | NAM-22 | North America | 310 | 260 | -33.99000 | 33.99000 | -28.51000 | 28.47000 | 0.2200 | 0.2200 | 83.00 | 42.50 |
| EUR-22 | 4 | EUR-25 | Europe | EUR-22 | Europe | 212 | 206 | -28.32000 | 18.10000 | -23.32000 | 21.78000 | 0.2200 | 0.2200 | -162.00 | 39.25 |
| AFR-22 | 5 | AFR-25 | Africa | AFR-22 | Africa | 388 | 402 | -24.75000 | 60.39000 | -45.87000 | 42.35000 | 0.2200 | 0.2200 | 180.00 | 90.00 |
| WAS-22 | 6 | WAS-25 | South Asia | WAS-22 | South Asia | 386 | 260 | -32.23000 | 52.47000 | -21.67000 | 35.31000 | 0.2200 | 0.2200 | -123.34 | 79.95 |
| EAS-22 | 7 | EAS-25 | East Asia | EAS-22 | East Asia | 396 | 251 | -43.23000 | 43.67000 | -22.10000 | 32.90000 | 0.2200 | 0.2200 | -64.78 | 77.61 |
| CAS-22 | 8 | CAS-25 | Central Asia | CAS-22 | Central Asia | 306 | 200 | -34.43000 | 32.67000 | -20.79000 | 22.99000 | 0.2200 | 0.2200 | -103.39 | 43.48 |
| AUS-22 | 9 | AUS-25 | Australasia | AUS-22 | Australasia | 400 | 258 | 142.05000 | 229.83000 | -22.99000 | 33.55000 | 0.2200 | 0.2200 | 141.38 | 60.31 |
| SEA-22 | 14 | SEA-25 | South East Asia | SEA-22 | South East Asia | 264 | 194 | 89.26000 | 147.12000 | -15.18000 | 27.28000 | 0.2200 | 0.2200 | 180.00 | 90.00 |
| SAM-44i | 1 | SAM-50i | South America | SAM-44i | South America | 181 | 155 | -106.25000 | -16.25000 | -58.25000 | 18.75000 | 0.5000 | 0.5000 | NaN | NaN |
| CAM-44i | 2 | CAM-50i | Central America | CAM-44i | Central America | 207 | 111 | -124.75000 | -21.75000 | -19.75000 | 35.25000 | 0.5000 | 0.5000 | NaN | NaN |
| NAM-44i | 3 | NAM-50i | North America | NAM-44i | North America | 300 | 129 | -171.75000 | -22.25000 | 12.25000 | 76.25000 | 0.5000 | 0.5000 | NaN | NaN |
| EUR-44i | 4 | EUR-50i | Europe | EUR-44i | Europe | 221 | 103 | -44.75000 | 65.25000 | 21.75000 | 72.75000 | 0.5000 | 0.5000 | NaN | NaN |
| AFR-44i | 5 | AFR-50i | Africa | AFR-44i | Africa | 173 | 179 | -25.25000 | 60.75000 | -46.25000 | 42.75000 | 0.5000 | 0.5000 | NaN | NaN |
| WAS-44i | 6 | WAS-50i | South Asia | WAS-44i | South Asia | 195 | 124 | 19.25000 | 116.25000 | -15.75000 | 45.75000 | 0.5000 | 0.5000 | NaN | NaN |
| EAS-44i | 7 | EAS-50i | East Asia | EAS-44i | East Asia | 227 | 157 | 62.75000 | 175.75000 | -18.75000 | 59.25000 | 0.5000 | 0.5000 | NaN | NaN |
| CAS-44i | 8 | CAS-50i | Central Asia | CAS-44i | Central Asia | 260 | 133 | 10.75000 | 140.25000 | 17.75000 | 69.75000 | 0.5000 | 0.5000 | NaN | NaN |
| AUS-44i | 9 | AUS-50i | Australasia | AUS-44i | Australasia | 238 | 133 | 88.75000 | 207.25000 | -53.25000 | 12.75000 | 0.5000 | 0.5000 | NaN | NaN |
| ANT-44i | 10 | ANT-50i | Antarctica | ANT-44i | Antarctica | 720 | 70 | -179.75000 | 179.75000 | -89.75000 | -55.25000 | 0.5000 | 0.5000 | NaN | NaN |
| ARC-44i | 11 | ARC-50i | Arctic | ARC-44i | Arctic | 720 | 83 | -179.75000 | 179.75000 | 48.75000 | 89.75000 | 0.5000 | 0.5000 | NaN | NaN |
| MED-44i | 12 | MED-50i | Mediterranean | MED-44i | Mediterranean | 144 | 65 | -20.75000 | 51.75000 | 25.25000 | 57.25000 | 0.5000 | 0.5000 | NaN | NaN |
| MNA-44i | 13 | MNA-50i | Middle East and North Africa | MNA-44i | Middle East and North Africa | 206 | 106 | -26.75000 | 75.75000 | -7.25000 | 45.25000 | 0.5000 | 0.5000 | NaN | NaN |
| MNA-22i | 13 | MNA-25i | Middle East and North Africa high res. | MNA-22i | Middle East and North Africa high res. | 410 | 209 | -26.62500 | 75.62500 | -6.87500 | 45.12500 | 0.2500 | 0.2500 | NaN | NaN |
| EUR-11i | 4 | EUR-12i | Europe high res. | EUR-11i | Europe high res. | 881 | 408 | -44.81250 | 65.18750 | 21.81250 | 72.68750 | 0.1250 | 0.1250 | NaN | NaN |
| SEA-22i | 14 | SEA-25i | South East Asia | SEA-22i | South East Asia | 233 | 172 | 89.12500 | 147.12500 | -15.37500 | 27.37500 | 0.2500 | 0.2500 | NaN | NaN |
EUR-11 example#
The heart of the module are some functions that create a dataset from the grid information, e.g.
eur11 = cx.cordex_domain("EUR-11", dummy="topo")
eur11
/Users/runner/micromamba/envs/cordex-tutorials/lib/python3.10/site-packages/cf_xarray/accessor.py:718: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
unused_keys = set(attribute.keys()) - set(inverted)
/Users/runner/micromamba/envs/cordex-tutorials/lib/python3.10/site-packages/cf_xarray/accessor.py:719: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
for key, value in attribute.items():
<xarray.Dataset> Size: 4MB
Dimensions: (rlon: 424, rlat: 412)
Coordinates:
* rlon (rlon) float64 3kB -28.38 -28.27 ... 18.05 18.16
* rlat (rlat) float64 3kB -23.38 -23.27 ... 21.73 21.84
lon (rlat, rlon) float64 1MB -10.06 -9.964 ... 64.96
lat (rlat, rlon) float64 1MB 21.99 22.03 ... 66.69
Data variables:
rotated_latitude_longitude int32 4B 0
topo (rlat, rlon) float32 699kB 284.0 246.0 ... 509.0
Attributes:
CORDEX_domain: EUR-11The dummy='topo' argument means, we want a dummy variable in the dataset to see how the domain looks like. For the dummy topography, we use the cdo topo operator in the background. So maybe you have to install python-cdo, e.g., conda install -c conda-forge python-cdo. Working with xarray datasets means, that we can use all the nice functions of xarray including plotting. The latest versions of py-cordex also come with an xarray accessor that allows you, e.g., to get a quick overview of the domain, using, e.g.
# from matplotlib import pyplot as plt
# plt.figure(figsize=(8,8))
eur11.cx.map()
Matplotlib is building the font cache; this may take a moment.
/Users/runner/micromamba/envs/cordex-tutorials/lib/python3.10/site-packages/cartopy/mpl/feature_artist.py:144: UserWarning: facecolor will have no effect as it has been defined as "never".
warnings.warn('facecolor will have no effect as it has been '
<GeoAxes: >
/Users/runner/micromamba/envs/cordex-tutorials/lib/python3.10/site-packages/cartopy/io/__init__.py:241: DownloadWarning: Downloading: https://naturalearth.s3.amazonaws.com/110m_physical/ne_110m_coastline.zip
warnings.warn(f'Downloading: {url}', DownloadWarning)
/Users/runner/micromamba/envs/cordex-tutorials/lib/python3.10/site-packages/cartopy/io/__init__.py:241: DownloadWarning: Downloading: https://naturalearth.s3.amazonaws.com/110m_cultural/ne_110m_admin_0_boundary_lines_land.zip
warnings.warn(f'Downloading: {url}', DownloadWarning)
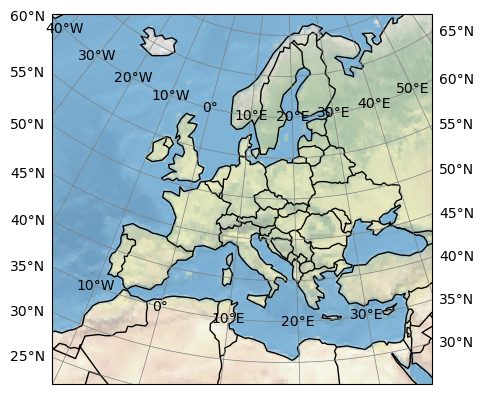
We can also use the dummy topography to plot an overview:
eur11.topo.plot(cmap="terrain")
<matplotlib.collections.QuadMesh at 0x11e46afe0>
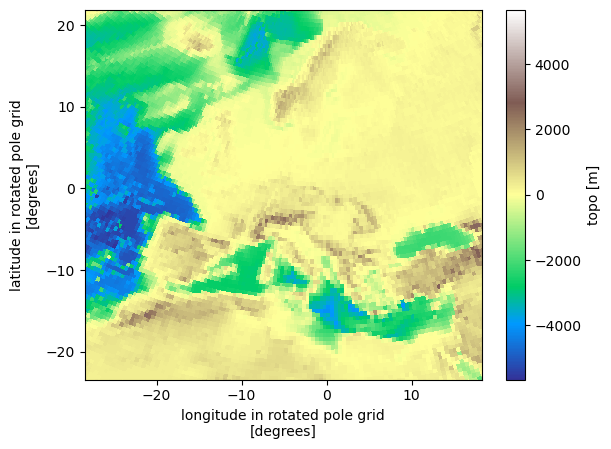
eur11.topo.plot(x="lon", y="lat", cmap="terrain")
<matplotlib.collections.QuadMesh at 0x11e51f820>
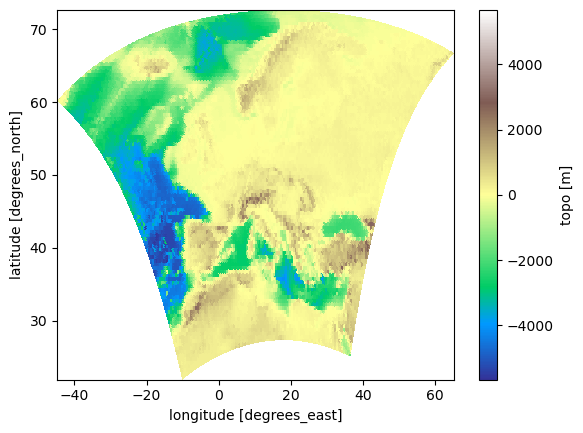
Let’s define a slightly more sophisticated plotting function that uses cartopy for the right projection with a rotated pole:
def plot(da, pole, vmin=None, vmax=None, borders=True, title=None):
"""plot a domain using the right projection with cartopy"""
import cartopy.crs as ccrs
import cartopy.feature as cf
import matplotlib.pyplot as plt
plt.figure(figsize=(10, 10))
projection = ccrs.PlateCarree()
transform = ccrs.RotatedPole(pole_latitude=pole[1], pole_longitude=pole[0])
# ax = plt.axes(projection=projection)
ax = plt.axes(projection=transform)
# ax.set_extent([ds_sub.rlon.min(), ds_sub.rlon.max(), ds_sub.rlat.min(), ds_sub.rlat.max()], crs=transform)
ax.gridlines(
draw_labels=True,
linewidth=0.5,
color="gray",
xlocs=range(-180, 180, 10),
ylocs=range(-90, 90, 5),
)
da.plot(
ax=ax,
cmap="terrain",
transform=transform,
vmin=vmin,
vmax=vmax,
x="rlon",
y="rlat",
)
ax.coastlines(resolution="50m", color="black", linewidth=1)
if borders:
ax.add_feature(cf.BORDERS)
if title is not None:
ax.set_title(title)
pole = (
eur11.rotated_latitude_longitude.grid_north_pole_longitude,
eur11.rotated_latitude_longitude.grid_north_pole_latitude,
)
pole
(-162.0, 39.25)
plot(eur11.topo, pole)
/Users/runner/micromamba/envs/cordex-tutorials/lib/python3.10/site-packages/cartopy/io/__init__.py:241: DownloadWarning: Downloading: https://naturalearth.s3.amazonaws.com/50m_physical/ne_50m_coastline.zip
warnings.warn(f'Downloading: {url}', DownloadWarning)
User defined domain#
The domains are actually created from a csv table. The domains are created using the create_dataset function, in which the data from the table is fed, e.g.:
cx.create_dataset?
Let’s create the EUR-11 domain manually from the numbers in the table:
cx.domains.table.loc["EUR-11"]
region 4
domain_id EUR-12
domain Europe
CORDEX_domain EUR-11
long_name Europe
nlon 424
nlat 412
ll_lon -28.375
ur_lon 18.155
ll_lat -23.375
ur_lat 21.835
dlon 0.11
dlat 0.11
pollon -162.0
pollat 39.25
Name: EUR-11, dtype: object
eur11_user = cx.create_dataset(
nlon=424,
nlat=412,
dlon=0.11,
dlat=0.11,
ll_lon=-28.375,
ll_lat=-23.375,
pollon=-162.00,
pollat=39.25,
dummy="topo",
)
/Users/runner/micromamba/envs/cordex-tutorials/lib/python3.10/site-packages/cf_xarray/accessor.py:718: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
unused_keys = set(attribute.keys()) - set(inverted)
/Users/runner/micromamba/envs/cordex-tutorials/lib/python3.10/site-packages/cf_xarray/accessor.py:719: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
for key, value in attribute.items():
We can check that this gives the same result as our preconfigured domain.
eur11_user.equals(eur11)
True
You can now use the create_dataset function to create any domain as an xarray dataset.
Plot all cordex-core domains#
We need a slightly modified plotting routine for this:
def plots(dsets, vmin=None, vmax=None, borders=True, title=None):
"""plot a domain using the right projection with cartopy"""
import cartopy.crs as ccrs
import cartopy.feature as cf
import matplotlib.patheffects as pe
import matplotlib.pyplot as plt
plt.figure(figsize=(20, 10))
projection = ccrs.PlateCarree()
# transform = ccrs.RotatedPole(pole_latitude=pole[1], pole_longitude=pole[0])
# ax = plt.axes(projection=projection)
ax = plt.axes(projection=projection)
# ax.set_extent([ds_sub.rlon.min(), ds_sub.rlon.max(), ds_sub.rlat.min(), ds_sub.rlat.max()], crs=transform)
ax.gridlines(
draw_labels=True,
linewidth=0.5,
color="gray",
xlocs=range(-180, 180, 15),
ylocs=range(-90, 90, 10),
)
# path_effects = [pe.Stroke(linewidth=50, foreground='g'), pe.Normal()]
for ds in dsets:
pole = (
ds.rotated_latitude_longitude.grid_north_pole_longitude,
ds.rotated_latitude_longitude.grid_north_pole_latitude,
)
transform = ccrs.RotatedPole(pole_latitude=pole[1], pole_longitude=pole[0])
ds.topo.plot(
ax=ax,
cmap="terrain",
transform=transform,
vmin=vmin,
vmax=vmax,
x="rlon",
y="rlat",
add_colorbar=False,
)
ax.coastlines(resolution="50m", color="black", linewidth=1)
if borders:
ax.add_feature(cf.BORDERS)
if title is not None:
ax.set_title("")
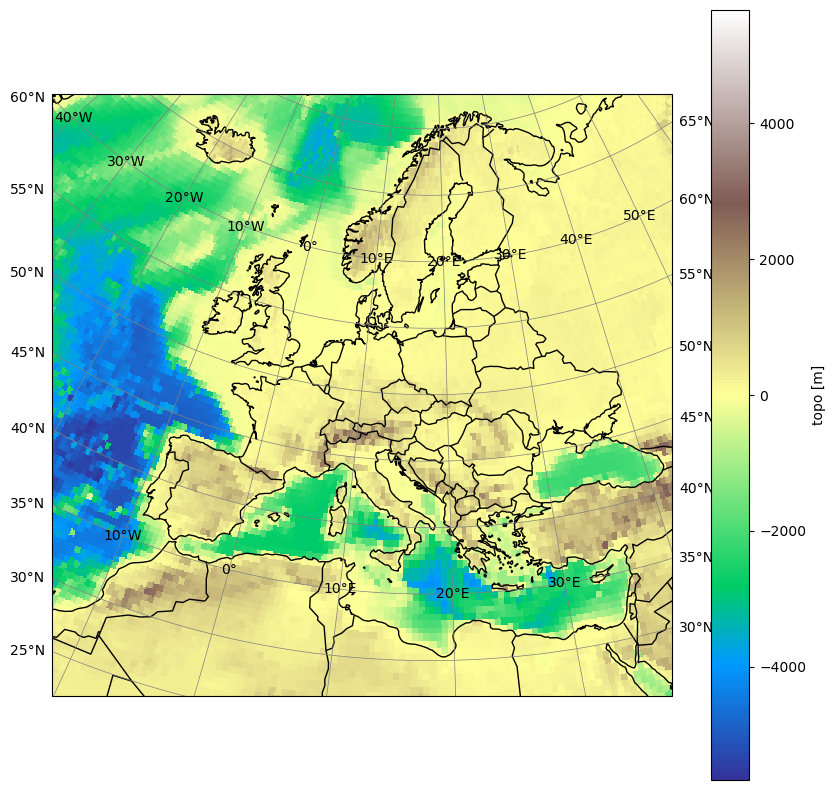
Now, let’s plot all cordex core domains into one overview. We will identify them by their resolution:
table = cx.domains.table
plots(
[
cx.cordex_domain(name, dummy="topo")
for name in table.index
if table.loc[name].dlon == 0.22
],
borders=False,
)
/Users/runner/micromamba/envs/cordex-tutorials/lib/python3.10/site-packages/cf_xarray/accessor.py:718: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
unused_keys = set(attribute.keys()) - set(inverted)
/Users/runner/micromamba/envs/cordex-tutorials/lib/python3.10/site-packages/cf_xarray/accessor.py:719: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
for key, value in attribute.items():
/Users/runner/micromamba/envs/cordex-tutorials/lib/python3.10/site-packages/cf_xarray/accessor.py:718: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
unused_keys = set(attribute.keys()) - set(inverted)
/Users/runner/micromamba/envs/cordex-tutorials/lib/python3.10/site-packages/cf_xarray/accessor.py:719: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
for key, value in attribute.items():
/Users/runner/micromamba/envs/cordex-tutorials/lib/python3.10/site-packages/cf_xarray/accessor.py:718: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
unused_keys = set(attribute.keys()) - set(inverted)
/Users/runner/micromamba/envs/cordex-tutorials/lib/python3.10/site-packages/cf_xarray/accessor.py:719: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
for key, value in attribute.items():
/Users/runner/micromamba/envs/cordex-tutorials/lib/python3.10/site-packages/cf_xarray/accessor.py:718: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
unused_keys = set(attribute.keys()) - set(inverted)
/Users/runner/micromamba/envs/cordex-tutorials/lib/python3.10/site-packages/cf_xarray/accessor.py:719: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
for key, value in attribute.items():
/Users/runner/micromamba/envs/cordex-tutorials/lib/python3.10/site-packages/cf_xarray/accessor.py:718: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
unused_keys = set(attribute.keys()) - set(inverted)
/Users/runner/micromamba/envs/cordex-tutorials/lib/python3.10/site-packages/cf_xarray/accessor.py:719: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
for key, value in attribute.items():
/Users/runner/micromamba/envs/cordex-tutorials/lib/python3.10/site-packages/cf_xarray/accessor.py:718: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
unused_keys = set(attribute.keys()) - set(inverted)
/Users/runner/micromamba/envs/cordex-tutorials/lib/python3.10/site-packages/cf_xarray/accessor.py:719: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
for key, value in attribute.items():
/Users/runner/micromamba/envs/cordex-tutorials/lib/python3.10/site-packages/cf_xarray/accessor.py:718: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
unused_keys = set(attribute.keys()) - set(inverted)
/Users/runner/micromamba/envs/cordex-tutorials/lib/python3.10/site-packages/cf_xarray/accessor.py:719: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
for key, value in attribute.items():
/Users/runner/micromamba/envs/cordex-tutorials/lib/python3.10/site-packages/cf_xarray/accessor.py:718: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
unused_keys = set(attribute.keys()) - set(inverted)
/Users/runner/micromamba/envs/cordex-tutorials/lib/python3.10/site-packages/cf_xarray/accessor.py:719: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
for key, value in attribute.items():
/Users/runner/micromamba/envs/cordex-tutorials/lib/python3.10/site-packages/cf_xarray/accessor.py:718: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
unused_keys = set(attribute.keys()) - set(inverted)
/Users/runner/micromamba/envs/cordex-tutorials/lib/python3.10/site-packages/cf_xarray/accessor.py:719: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
for key, value in attribute.items():
/Users/runner/micromamba/envs/cordex-tutorials/lib/python3.10/site-packages/cf_xarray/accessor.py:718: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
unused_keys = set(attribute.keys()) - set(inverted)
/Users/runner/micromamba/envs/cordex-tutorials/lib/python3.10/site-packages/cf_xarray/accessor.py:719: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
for key, value in attribute.items():
/Users/runner/micromamba/envs/cordex-tutorials/lib/python3.10/site-packages/cf_xarray/accessor.py:718: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
unused_keys = set(attribute.keys()) - set(inverted)
/Users/runner/micromamba/envs/cordex-tutorials/lib/python3.10/site-packages/cf_xarray/accessor.py:719: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
for key, value in attribute.items():
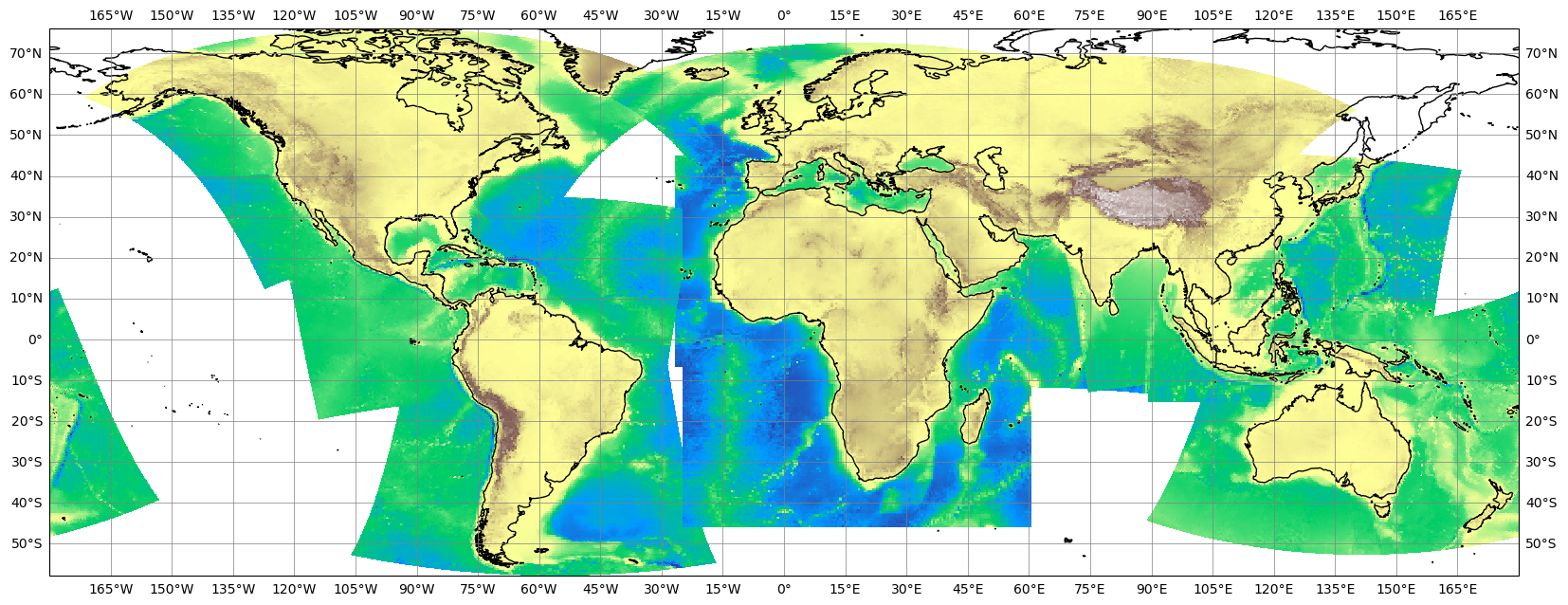
Using the grid information for interpolation#
The gridded Cordex datasets are particular usefule for regridding either with CDO or other interpolation packages.
We will use some CMIP6 model data from the ESGF to show how we can do the regridding:
!wget https://raw.githubusercontent.com/remo-rcm/pyremo-data/main/orog_fx_MPI-ESM1-2-HR_historical_r1i1p1f1_gn.nc
--2025-04-01 21:35:15-- https://raw.githubusercontent.com/remo-rcm/pyremo-data/main/orog_fx_MPI-ESM1-2-HR_historical_r1i1p1f1_gn.nc
Resolving raw.githubusercontent.com (raw.githubusercontent.com)... 185.199.111.133, 185.199.109.133, 185.199.110.133, ...
Connecting to raw.githubusercontent.com (raw.githubusercontent.com)|185.199.111.133|:443... connected.
HTTP request sent, awaiting response...
200 OK
Length: 299137 (292K)
[application/octet-stream]
Saving to: ‘orog_fx_MPI-ESM1-2-HR_historical_r1i1p1f1_gn.nc.1’
orog_fx_M 0%[ ] 0 --.-KB/s
orog_fx_MPI-ESM1-2- 100%[===================>] 292.13K --.-KB/s in 0.02s
2025-04-01 21:35:15 (11.9 MB/s) - ‘orog_fx_MPI-ESM1-2-HR_historical_r1i1p1f1_gn.nc.1’ saved [299137/299137]
import xarray as xr
# https://raw.githubusercontent.com/remo-rcm/pyremo-data/main/orog_fx_MPI-ESM1-2-HR_historical_r1i1p1f1_gn.nc
ds = xr.open_dataset("orog_fx_MPI-ESM1-2-HR_historical_r1i1p1f1_gn.nc")
# ds = xr.open_dataset(
# "https://esgf3.dkrz.de/thredds/dodsC/cmip6/CMIP/MPI-M/MPI-ESM1-2-HR/historical/r1i1p1f1/Amon/tas/gn/v20190710/tas_Amon_MPI-ESM1-2-HR_historical_r1i1p1f1_gn_199501-199912.nc"
# )
# ds = xr.open_dataset(
# "http://esgf-data3.ceda.ac.uk/thredds/dodsC/esg_cmip6/CMIP6/CMIP/MOHC/UKESM1-0-LL/historical/r1i1p1f2/Amon/tas/gn/v20190406/tas_Amon_UKESM1-0-LL_historical_r1i1p1f2_gn_185001-194912.nc"
# )
ds
<xarray.Dataset> Size: 309kB
Dimensions: (lat: 192, bnds: 2, lon: 384)
Coordinates:
* lat (lat) float64 2kB -89.28 -88.36 -87.42 ... 87.42 88.36 89.28
* lon (lon) float64 3kB 0.0 0.9375 1.875 2.812 ... 357.2 358.1 359.1
Dimensions without coordinates: bnds
Data variables:
lat_bnds (lat, bnds) float64 3kB ...
lon_bnds (lon, bnds) float64 6kB ...
orog (lat, lon) float32 295kB ...
Attributes: (12/47)
Conventions: CF-1.7 CMIP-6.2
activity_id: CMIP
branch_method: standard
branch_time_in_child: 0.0
branch_time_in_parent: 0.0
contact: cmip6-mpi-esm@dkrz.de
... ...
title: MPI-ESM1-2-HR output prepared for CMIP6
tracking_id: hdl:21.14100/3341d289-1282-489f-9419-c263718da247
variable_id: orog
variant_label: r1i1p1f1
license: CMIP6 model data produced by MPI-M is licensed un...
cmor_version: 3.5.0Interpolation using CDO#
We will use CDO’s python bindings to control the cdo operators. Please note, that the python bindings in the background actually execute the cdo commands on the command line. CDO does have a huge IO overhead since it will always write a file between each operator step and will always need data from a file on the filesystem as input. If you give an xarray dataset as input (see below), the python binding will actually trigger a to_netcdf call to write the input as a temporary file to disk. You should be aware of this if you use huge xarray datasets as input.
We will first write the EUR-11 grid into a file on the disk so that we can use it as input to CDO
from cdo import Cdo
eur11.to_netcdf("EUR-11_grid.nc")
Now we will remap the first timestep of the CMIP6 modeldata to the EUR-11 grid:
remap_cdo = Cdo().remapbil("EUR-11_grid.nc", input=ds, returnXArray="orog")
remap_cdo
<xarray.DataArray 'orog' (rlat: 412, rlon: 424)> Size: 699kB
[174688 values with dtype=float32]
Coordinates:
lon (rlat, rlon) float64 1MB ...
lat (rlat, rlon) float64 1MB ...
* rlon (rlon) float64 3kB -28.38 -28.27 -28.16 ... 17.93 18.05 18.16
* rlat (rlat) float64 3kB -23.38 -23.27 -23.16 ... 21.61 21.73 21.84
Attributes:
standard_name: surface_altitude
long_name: Surface Altitude
units: m
grid_mapping: rotated_latitude_longitude
comment: The surface called 'surface' means the lower boundary of ...
original_name: orog
cell_methods: area: mean
cell_measures: area: areacella
history: 2019-08-25T06:42:14Z altered by CMOR: replaced missing va...remap_cdo.plot()
<matplotlib.collections.QuadMesh at 0x124328bb0>
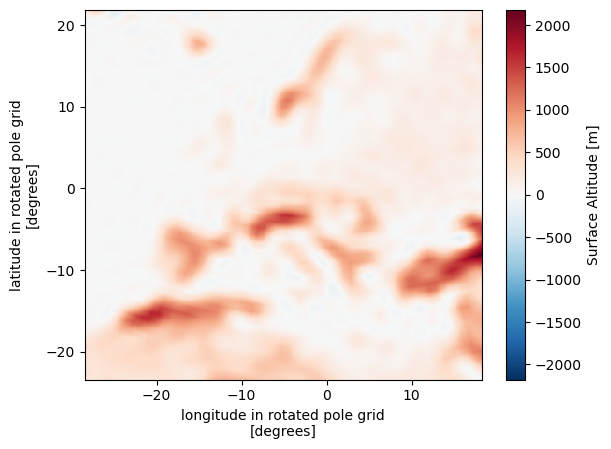
Alternative interpolation methods#
A nice alternative is xesmf since it is based on xarray and will also very nicely work with dask.
import xesmf as xe
regridder = xe.Regridder(ds, eur11, "bilinear", periodic=True)
regridder
xESMF Regridder
Regridding algorithm: bilinear
Weight filename: bilinear_192x384_412x424_peri.nc
Reuse pre-computed weights? False
Input grid shape: (np.int32(192), np.int32(384))
Output grid shape: (np.int32(412), np.int32(424))
Periodic in longitude? True
remap_xe = regridder(ds.orog)
remap_xe.plot()
<matplotlib.collections.QuadMesh at 0x136677370>
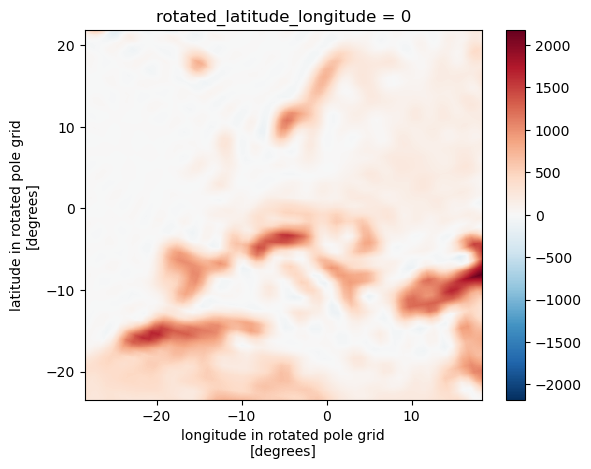
We can easily compare both approaches
(remap_cdo - remap_xe).plot(x="lon", y="lat")
<matplotlib.collections.QuadMesh at 0x1366fb0d0>
The nice thing about xesmf is that it works together with xarray and will keep all meta information. Another consequence is that xesmf works well with dask and it’s vectorization. That means, if we have a long time axis along we want to regrid, this can easily be parallelized using, e.g., dask.distributed and will also work nicely with large datasets that don’t fit into memory. The xesmf regridder can also store and reuse regridding weights for faster interpolation.

